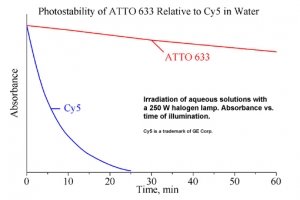
ATTO 633







|
Optische Eigenschaften |
||||
|
λAabs |
630 |
nm |
||
|
εmax |
1,3×105 |
M-1 cm-1 |
||
|
λfl |
651 |
nm |
||
|
ηfl |
64 |
% |
||
|
τfl |
3,3 |
ns |
||
|
CF260 = ε260/εmax |
0,04 |
|
||
|
CF280 = ε280/εmax |
0,05 |
|
||
ATTO 633 zählt im roten Spektralbereich zu einer neuen – speziell von ATTO-TEC entwickelten – Generation von Fluoreszenzmarkern. Zu den charakteristischen Eigenschaften des Farbstoffs zählen starke Absorption, sehr hohe Fluoreszenzquantenausbeute sowie hohe thermische und photochemische Stabilität. ATTO 633 ist somit besonders geeignet im Bereich der Einzelmoleküldetektion sowie zur hochauflösenden Mikroskopie (SIM, STED, etc.).
Die Absorption und Fluoreszenz ist im pH-Bereich von 2 - 11 pH-unabhängig. Der Farbstoff ist mäßig hydrophil. ATTO 633 ist ein kationischer Farbstoff. Nach der Kopplung an ein Substrat ist der Farbstoff einfach positiv geladen. Eine effiziente Anregung der Fluoreszenz erfolgt im Bereich 610 - 645 nm. Eine für ATTO 633 geeignete Anregungsquelle ist die 633 nm Linie des HeNe-Lasers sowie Diodenlaser mit Emission bei 635 nm.
Produktdatenblätter
- ATTO 633 Produktinformation
- NHS-Ester
- Maleimid
- Iodacetamid
- Phalloidin
- Amin
- Azid/Alkin
- Streptavidin
Absorptions- und Emissionsspektrum (ASCII)
- ATTO 633 Absorption (.txt)
- ATTO 633 Emission (.txt)
Absorptions- und Emissionsspektrum (Grafik)
- ATTO 633 Absorption/Fluoreszenz (.jpg)
Ausgewählte Literatur zu ATTO 633:
M. Bao, J. Xie, Geometric Confinement-Mediated Mechanical Tension Directs Patterned Differentiation of Mouse ESCs into Organized Germ Layers, ACS Applied Materials & Interfaces 15, 34397 (2023).
P. Dave, G. Roth, E. Griesbach, D. Mateju, T. Hochstoeger, J. Chao, Single-molecule imaging reveals translation-dependent destabilization of mRNAs, Molecular Cell 83, 589-606.e6 (2023).
A. Di Costagliola Polidoro, Z. Baghbantarghdari, V. de Gregorio, S. Silvestri, P. Netti, E. Torino, Insulin Activation Mediated by Uptake Mechanisms, Biomacromolecules 24, 2203 (2023).
T. Gui, C. Fleming, C. Manzato, B. Bourgeois, N. Sirati, J. Heuer, I. Papadionysiou, D. van Montfort, M. van Gijzen, L. Smits, B. Burgering, T. Madl, J. Schuijers, Targeted perturbation of signaling-driven condensates, Molecular Cell 83, 4141-4157.e11 (2023).
I. Hefetz, O. Israeli, G. Bilinsky, I. Plaschkes, E. Hazkani-Covo, Z. Hayouka, A. Lampert, Y. Helman, A reversible mutation in a genomic hotspot saves bacterial swarms from extinction, iScience 26, 106043 (2023).
X. Jiang, J. Mietner, J. Navarro, A combination of surface-initiated controlled radical polymerization (SET-LRP) and click-chemistry for the chemical modification and fluorescent labeling of cellulose nanofibrils, Cellulose 30, 2929 (2023).
V. Karema-Jokinen, A. Koskela, M. Hytti, H. Hongisto, T. Viheriälä, M. Liukkonen, T. Torsti, H. Skottman, A. Kauppinen, S. Nymark, K. Kaarniranta, Crosstalk of protein clearance, inflammasome, and Ca2+ channels in retinal pigment epithelium derived from age-related macular degeneration patients, Journal of Biological Chemistry 299, 104770 (2023).
E. Krok, H. Franquelim, M. Chattopadhyay, H. Orlikowska-Rzeznik, P. Schwille, L. Piatkowski, Nanoscale structural response of biomimetic cell membranes to controlled dehydration, Nanoscale 16, 72 (2023).
E. Krok, M. Stephan, R. Dimova, L. Piatkowski, Tunable biomimetic bacterial membranes from binary and ternary lipid mixtures and their application in antimicrobial testing, Biochimica et Biophysica Acta (BBA) - Biomembranes 1865, 184194 (2023).
E. Mäki-Mikola, P. Lauren, N. Uema, K. Kikuchi, Y. Takashima, T. Laaksonen, T. Lajunen, Establishing a simple perfusion cell culture system for light-activated liposomes, Scientific reports 13, 2050 (2023).
A. Mirza, M. Akhtar, J. Aguren, J. Marino, J. Bruno, Advancements in Rapid and Affordable Diagnostic Testing for Respiratory Infectious Diseases, Journal of fluorescence (2023).
P. Perez Schmidt, T. Luedtke, P. Moretti, P. Di Gianvincenzo, M. Fernandez Leyes, B. Espuche, H. Amenitsch, G. Wang, H. Ritacco, L. Polito, M. Ortore, S. Moya, Assembly and recognition mechanisms of glycosylated PEGylated polyallylamine phosphate nanoparticles, Journal of Colloid and Interface Science 645, 448 (2023).
K. Riedlová, M. Saija, A. Olżyńska, P. Jurkiewicz, P. Daull, J.-S. Garrigue, L. Cwiklik, Influence of BAKs on tear film lipid layer, European Journal of Pharmaceutics and Biopharmaceutics 186, 65 (2023).
R. Sahu, C. Goswami, Presence of TRPV3 in macrophage lysosomes helps in skin wound healing against bacterial infection, Experimental Dermatology 32, 60 (2023).
R. West, K. Kanellopulos, S. Schmid, Photothermal Microscopy and Spectroscopy with Nanomechanical Resonators, The Journal of Physical Chemistry C 127, 21915 (2023).
L. Cassella, A. Ephrussi, Subcellular spatial transcriptomics identifies three mechanistically different classes of localizing RNAs, Nature Communications 13, 1 (2022).
S. Dey, A. Dorey, L. Abraham, Y. Xing, I. Zhang, F. Zhang, S. Howorka, H. Yan, A reversibly gated protein-transporting membrane channel made of DNA, Nature Communications 13, 1 (2022).
J. Dodge, A. Doyle, A. Costa-da-Silva, C. Hogden, E. Mezey, J. Mays, Atto 465 Derivative Is a Nuclear Stain with Unique Excitation and Emission Spectra Useful for Multiplex Immunofluorescence Histochemistry, Journal of Histochemistry & Cytochemistry 70, 211 (2022).
V. Kast, A. Nadernezhad, D. Pette, A. Gabrielyan, M. Fusenig, K. Honselmann, D. Stange, C. Werner, D. Loessner, A Tumor Microenvironment Model of Pancreatic Cancer to Elucidate Responses toward Immunotherapy, Advanced Healthcare Materials n/a, 2201907 (2022).
M. Klein, K. Shulenberger, U. Barotov, T. Šverko, M. Bawendi, Supramolecular Lattice Deformation and Exciton Trapping in Nanotubular J-Aggregates, The Journal of Physical Chemistry C 126, 4095 (2022).
E. Krok, A. Batura, M. Chattopadhyay, H. Orlikowska, L. Piatkowski, Lateral organization of biomimetic cell membranes in varying pH conditions, Journal of Molecular Liquids 345, 117907 (2022).
N. Louros, M. Ramakers, E. Michiels, K. Konstantoulea, C. Morelli, T. Garcia, N. Moonen, S. D’Haeyer, V. Goossens, D. Thal, D. Audenaert, F. Rousseau, J. Schymkowitz, Mapping the sequence specificity of heterotypic amyloid interactions enables the identification of aggregation modifiers, Nature Communications 13, 1 (2022).
N. Mirzazadeh Dizaji, Y. Lin, T. Bein, E. Wagner, S. Wuttke, U. Lächelt, H. Engelke, Biomimetic Mineralization of Iron-Fumarate Nanoparticles for Protective Encapsulation and Intracellular Delivery of Proteins, Chemistry of materials : a publication of the American Chemical Society 34, 8684 (2022).
M. Nguyen, D. Biriukov, C. Tempra, K. Baxova, H. Martinez-Seara, H. Evci, V. Singh, R. Šachl, M. Hof, P. Jungwirth, M. Javanainen, M. Vazdar, Ionic Strength and Solution Composition Dictate the Adsorption of Cell-Penetrating Peptides onto Phosphatidylcholine Membranes, Langmuir : the ACS journal of surfaces and colloids 38, 11284 (2022).
N. Niranjan Dhanasekar, D. Thiyagarajan, D. Bhatia, DNA Origami in the Quest for Membrane Piercing, Chemistry – An Asian Journal 17, e202200591 (2022).
S. Pritzl, D. Konrad, M. Ober, A. Richter, J. Frank, B. Nickel, D. Trauner, T. Lohmüller, Optical Membrane Control with Red Light Enabled by Red-Shifted Photolipids, Langmuir 38, 385 (2022).
S. Pritzl, J. Morstein, S. Kahler, D. Konrad, D. Trauner, T. Lohmüller, Postsynthetic Photocontrol of Giant Liposomes via Fusion-Based Photolipid Doping, Langmuir : the ACS journal of surfaces and colloids 38, 11941 (2022).
P. Ramamurthi, Z. Zhao, E. Burke, N. Steinmetz, M. Müllner, Tuning the Hydrophilic–Hydrophobic Balance of Molecular Polymer Bottlebrushes Enhances their Tumor Homing Properties, Advanced Healthcare Materials 11, 2200163 (2022).
P. Sutthavas, Z. Tahmasebi Birgani, P. Habibovic, S. van Rijt, Calcium Phosphate-Coated and Strontium-Incorporated Mesoporous Silica Nanoparticles Can Effectively Induce Osteogenic Stem Cell Differentiation, Advanced Healthcare Materials 11, 2101588 (2022).
H. Varol, S. Seeger, Fluorescent Staining of Silicone Micro- and Nanopatterns for Their Optical Imaging, Langmuir 38, 231 (2022).
Z. Zhao, D. Roy, J. Steinkühler, T. Robinson, R. Lipowsky, R. Dimova, Super-Resolution Imaging of Highly Curved Membrane Structures in Giant Vesicles Encapsulating Molecular Condensates, Advanced Materials 34, 2106633 (2022).
T. Altman, A. Ionescu, A. Ibraheem, D. Priesmann, T. Gradus-Pery, L. Farberov, G. Alexandra, N. Shelestovich, R. Dafinca, N. Shomron, F. Rage, K. Talbot, M. Ward, A. Dori, M. Krüger, E. Perlson, Axonal TDP-43 condensates drive neuromuscular junction disruption through inhibition of local synthesis of nuclear encoded mitochondrial proteins, Nature Communications 12, 1 (2021).
J. Auerswald, J. Ebenhan, C. Schwieger, A. Scrima, A. Meister, K. Bacia, Measuring protein insertion areas in lipid monolayers by fluorescence correlation spectroscopy, Biophysical Journal 120 (2021).
B. Benham-Pyle, C. Brewster, A. Kent, F. Mann, S. Chen, A. Scott, A. Box, A. Sánchez Alvarado, Identification of rare, transient post-mitotic cell states that are induced by injury and required for whole-body regeneration in Schmidtea mediterranea, Nature Cell Biology 23, 939 (2021).
J. Burke, A. Gilchrist, S. Sawyer, R. Parker, RNase L limits host and viral protein synthesis via inhibition of mRNA export, Science Advances 7, 120 (2021).
M. Chattopadhyay, E. Krok, H. Orlikowska, P. Schwille, H. Franquelim, L. Piatkowski, Hydration Layer of Only a Few Molecules Controls Lipid Mobility in Biomimetic Membranes, Journal of the American Chemical Society 143, 14551 (2021).
M. Klein, K. Shulenberger, U. Barotov, T. Šverko, M. Bawendi, Supramolecular Lattice Deformation and Exciton Trapping in Nanotubular J-Aggregates, The Journal of Physical Chemistry C 126, 4095 (2022).
J. Lee, K. Lee, C. Wang, D. Ha, G.-H. Kim, J. Park, T. Kim, Combined Effects of Zeta-potential and Temperature of Nanopores on Diffusioosmotic Ion Transport, Analytical Chemistry 93, 14169 (2021).
R. Leisi, E. Widmer, B. Gooch, N. Roth, C. Ros, Mechanistic insights into flow-dependent virus retention in different nanofilter membranes, Journal of Membrane Science 636, 119548 (2021).
A. Niederberger, T. Pelras, L. Manni, P. FitzGerald, G. Warr, M. Müllner, Stiffness-Dependent Intracellular Location of Cylindrical Polymer Brushes, Macromolecular Rapid Communications 42, 2100138 (2021).
M. Partipilo, E. Ewins, J. Frallicciardi, T. Robinson, B. Poolman, D. Slotboom, Minimal Pathway for the Regeneration of Redox Cofactors, JACS Au 1, 2280 (2021).
S. Safarian, P. Kusov, S. Kosolobov, O. Borzenkova, A. Khakimov, Y. Kotelevtsev, V. Drachev, Surface-specific washing-free immunosensor for time-resolved cortisol monitoring, Talanta 225, 122070 (2021).
S. Suarasan, C. Tira, M. Rusu, A.-M. Craciun, M. Focsan, Controlled fluorescence manipulation by core-shell multilayer of spherical gold nanoparticles, Journal of Molecular Structure 1244, 130950 (2021).
H. Ucar, H.-A. Wagenknecht, DNA-templated control of chirality and efficient energy transport in supramolecular DNA architectures with aggregation-induced emission, Chemical Science 12, 10048 (2021).
J. Xie, M. Bao, X. Hu, W. Koopman, W. Huck, Energy expenditure during cell spreading influences the cellular response to matrix stiffness, Biomaterials 267, 120494 (2021).
Y. Xue, I. Braslavsky, S. Quake, Temperature effect on polymerase fidelity, Journal of Biological Chemistry 297, 101270 (2021).
A. Zengin, J. Castro, P. Habibovic, S. van Rijt, Injectable, self-healing mesoporous silica nanocomposite hydrogels with improved mechanical properties, Nanoscale 13, 1144 (2021).
S. Azinheiro, K. Kant, M.-A. Shahbazi, A. Garrido-Maestu, M. Prado, L. Dieguez, A smart microfluidic platform for rapid multiplexed detection of foodborne pathogens, Food Control 114, 107242 (2020).
D. Bölükbas, S. Datz, C. Meyer-Schwickerath, C. Morrone, A. Doryab, D. Gößl, M. Vreka, L. Yang, C. Argyo, S. van Rijt, M. Lindner, O. Eickelberg, T. Stoeger, O. Schmid, S. Lindstedt, G. Stathopoulos, T. Bein, D. Wagner, S. Meiners, Organ-Restricted Vascular Delivery of Nanoparticles for Lung Cancer Therapy, Advanced Therapeutics 3, 2000017 (2020).
D. Boytsov, C. Hannesschlaeger, A. Horner, C. Siligan, P. Pohl, Micropipette Aspiration-Based Assessment of Single Channel Water Permeability, Biotechnology Journal 15, 1900450 (2020).
L. Chen, X. Chen, X. Yang, C. He, M. Wang, P. Xi, J. Gao, Advances of super-resolution fluorescence polarization microscopy and its applications in life sciences, Computational and Structural Biotechnology Journal 18, 2209 (2020).
A. Milewska, K. Owczarek, A. Szczepanski, K. Pyrc, Visualizing Coronavirus Entry into Cells, Methods in molecular biology (Clifton, N.J.) 2203, 241 (2020).
A. Olżyńska, A. Wizert, M. Štefl, D. Iskander, L. Cwiklik, Mixed polar-nonpolar lipid films as minimalistic models of Tear Film Lipid Layer, Biochimica et Biophysica Acta (BBA) - Biomembranes 1862, 183300 (2020).
M. Reichardt, C. Neuhaus, J.-D. Nicolas, M. Bernhardt, K. Toischer, T. Salditt, X-Ray Structural Analysis of Single Adult Cardiomyocytes, Biophysical Journal 119, 1309 (2020).
M. Sabatino, F. Carfì Pavia, S. Rigogliuso, D. Giacomazza, G. Ghersi, V. La Carrubba, C. Dispenza, Development of injectable and durable kefiran hydro-alcoholic gels, International Journal of Biological Macromolecules 149, 309 (2020).
M. Sabatino, L. Ditta, A. Conigliaro, C. Dispenza, A multifuctional nanoplatform for drug targeted delivery based on radiation-engineered nanogels, Radiation Physics and Chemistry 169, 108059 (2020).
R. Šachl, S. Čujová, V. Singh, P. Riegerová, P. Kapusta, H.-M. Müller, J. Steringer, M. Hof, W. Nickel, Functional Assay to Correlate Protein Oligomerization States with Membrane Pore Formation, Analytical Chemistry 92, 14861 (2020).
J. Hu, S. Gupta, J. Ozdemir, M. Beyzavi, Applications of Dynamic Covalent Chemistry Concept toward Tailored Covalent Organic Framework Nanomaterials, ACS Applied Nano Materials 3, 6239 (2020).
N. Larkey, J. Phillips, H. Jang, S. Kolluri, S. Burrows, Small RNA Biosensor Design Strategy To Mitigate Off-Analyte Response, ACS Sensors 5, 377 (2020).
L. Andrée, D. Barata, P. Sutthavas, P. Habibovic, S. van Rijt, Guiding mesenchymal stem cell differentiation using mesoporous silica nanoparticle-based films, Acta Biomaterialia 96, 557 (2019).
N. Garino, T. Limongi, B. Dumontel, M. Canta, L. Racca, M. Laurenti, M. Castellino, A. Casu, A. Falqui, V. Cauda, A Microwave-Assisted Synthesis of Zinc Oxide Nanocrystals Finely Tuned for Biological Applications, Nanomaterials 9, 212 (2019).
T. Godjevargova, Y. Ivanov, M. Atanasova, Z. Becheva, A. Zherdev, Magnetic nanoparticles based fluorescence immunoassay for food contaminants, Food Science and Applied Biotechnology 2, 38 (2019).
B. Hampoelz, A. Schwarz, P. Ronchi, H. Bragulat-Teixidor, C. Tischer, I. Gaspar, A. Ephrussi, Y. Schwab, M. Beck, Nuclear Pores Assemble from Nucleoporin Condensates During Oogenesis, Cell 179, 671-686.e17 (2019).
J.-D. Nicolas, M. Bernhardt, S. Schlick, M. Tiburcy, W.-H. Zimmermann, A. Khan, A. Markus, F. Alves, K. Toischer, T. Salditt, X-ray diffraction imaging of cardiac cells and tissue, Progress in Biophysics and Molecular Biology 144, 151 (2019).
P. Picone, M. Sabatino, A. Ajovalasit, D. Giacomazza, C. Dispenza, M. Di Carlo, Biocompatibility, hemocompatibility and antimicrobial properties of xyloglucan-based hydrogel film for wound healing application, International Journal of Biological Macromolecules 121, 784 (2019).
B. Ramesh, S. Abnouf, S. Mali, W. Moree, U. Patil, S. Bark, N. Varadarajan, Engineered ChymotrypsiN for Mass Spectrometry-Based Detection of Protein Glycosylation, ACS Chemical Biology 14, 2616 (2019).
K. Sy Piecco, J. Vicente, J. Pyle, D. Ingram, M. Kordesch, J. Chen, Reusable Chemically Micropatterned Substrates via Sequential Photoinitiated Thiol–Ene Reactions as a Template for Perovskite Thin-Film Microarrays, ACS Applied Electronic Materials 1, 2279 (2019).
R. Thomsen, M. Galsgaard Malle, A. Hauge Okholm, S. Krishnan, S. Bohr, R. Schøler Sørensen, O. Ries, S. Vogel, F. Simmel, N. Hatzakis, J. Kjems, A large size-selective DNA nanopore with sensing applications, Nature Communications 10, 1 (2019).
S. Wagle, V. Georgiev, T. Robinson, R. Dimova, R. Lipowsky, A. Grafmüller, Interaction of SNARE Mimetic Peptides with Lipid bilayers, Scientific Reports 9, 7708 (2019).
O. Wahlsten, F. Ulander, D. Midtvedt, M. Henningson, V. Zhdanov, B. Agnarsson, F. Höök, Quantitative Detection of Biological Nanoparticles in Solution via Their Mediation of Colocalization of Fluorescent Liposomes, Physical Review Applied 12 (2019).
P. Chidchob, D. Offenbartl-Stiegert, D. McCarthy, X. Luo, J. Li, S. Howorka, H. Sleiman, Spatial Presentation of Cholesterol Units on a DNA Cube as a Determinant of Membrane Protein-Mimicking Functions, Journal of the American Chemical Society 141, 1100 (2019).
S. Krause, T. Vosch, Stokes shift microscopy by excitation and emission imaging, Optics Express 27, 8208 (2019).
H. Xiong, N. Qian, Y. Miao, Z. Zhao, W. Min, Stimulated Raman Excited Fluorescence Spectroscopy of Visible Dyes, The Journal of Physical Chemistry Letters 10, 3563 (2019).
L. Avila, R. Chandrasekar, K. Wilkinson, J. Balthazor, M. Heerman, J. Bechard, S. Brown, Y. Park, G. Reek, J. Tomich, Delivery of lethal dsRNAs in insect diets by branched amphiphilic peptide capsules, Journal of Controlled Release 273, 139 (2018).
M. Bernhardt, J.-D. Nicolas, M. Osterhoff, H. Mittelstädt, M. Reuss, B. Harke, A. Wittmeier, M. Sprung, S. Köster, T. Salditt, Correlative microscopy approach for biology using X-ray holography, X-ray scanning diffraction and STED microscopy, Nature Communications 9, 3641 (2018).
E. Braselmann, A. Wierzba, J. Polaski, M. Chromiński, Z. Holmes, S.-T. Hung, D. Batan, J. Wheeler, R. Parker, R. Jimenez, D. Gryko, R. Batey, A. Palmer, A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells, Nature chemical biology 14, 964 (2018).
F. Fenaroli, U. Repnik, Y. Xu, K. Johann, S. van Herck, P. Dey, F. Skjeldal, D. Frei, S. Bagherifam, A. Kocere, R. Haag, B. de Geest, M. Barz, D. Russell, G. Griffiths, Enhanced Permeability and Retention-like Extravasation of Nanoparticles from the Vasculature into Tuberculosis Granulomas in Zebrafish and Mouse Models, ACS Nano 12, 8646 (2018).
E. Horak, R. Goldsmith, Drumming up single-molecule beats, Proceedings of the National Academy of Sciences 115, 11115 (2018).
S. Krause, M. Koerstz, R. Arppe-Tabbara, T. Soukka, T. Vosch, NIR induced modulation of the red emission from erbium ions for selective lanthanide imaging, Methods and Applications in Fluorescence 6, 44001 (2018).
T. Sunami, K. Shimada, G. Tsuji, S. Fujii, Flow Cytometric Analysis To Evaluate Morphological Changes in Giant Liposomes As Observed in Electrofusion Experiments, Langmuir 34, 88 (2018).
W. Xiang, M. Roberti, J.-K. Hériché, S. Huet, S. Alexander, J. Ellenberg, Correlative live and super-resolution imaging reveals the dynamic structure of replication domains, The Journal of Cell Biology 217, 1973 (2018).
N. Burma, R. Bonin, H. Leduc-Pessah, C. Baimel, Z. Cairncross, M. Mousseau, J. Shankara, P. Stemkowski, D. Baimoukhametova, J. Bains, M. Antle, G. Zamponi, C. Cahill, S. Borgland, Y. de Koninck, T. Trang, Blocking microglial pannexin-1 channels alleviates morphine withdrawal in rodents, Nature medicine 23, 355 (2017).
M. Jbeily, J. Kressler, Fluorophilicity and lipophilicity of fluorinated rhodamines determined by their partition coefficients in biphasic solvent systems, Journal of Fluorine Chemistry 193, 67 (2017).
M. Santi, G. Maccari, P. Mereghetti, V. Voliani, S. Rocchiccioli, N. Ucciferri, S. Luin, G. Signore, Rational Design of a Transferrin-Binding Peptide Sequence Tailored to Targeted Nanoparticle Internalization, Bioconjugate Chemistry 28, 471 (2017).
K. Grußmayer, D.-P. Herten, Time-resolved molecule counting by photon statistics across the visible spectrum, Physical chemistry chemical physics : PCCP 19, 8962 (2017).
J. Hackett, X. Shi, A. Kobylarz, M. Lucas, R. Wessendorf, K. Hines, S. Bentolila, M. Hanson, Y. Lu, An Organelle RNA Recognition Motif Protein Is Required for Photosystem II Subunit psbF Transcript Editing, Plant physiology 173, 2278 (2017).
H. Leduc-Pessah, N. Weilinger, C. Fan, N. Burma, R. Thompson, T. Trang, Site-Specific Regulation of P2X7 Receptor Function in Microglia Gates Morphine Analgesic Tolerance, The Journal of neuroscience : the official journal of the Society for Neuroscience 37, 10154 (2017).
M. Lunova, A. Prokhorov, M. Jirsa, M. Hof, A. Olżyńska, P. Jurkiewicz, Š. Kubinová, O. Lunov, A. Dejneka, Nanoparticle core stability and surface functionalization drive the mTOR signaling pathway in hepatocellular cell lines, Scientific Reports 7, 16049 (2017).
M. Mahmud-Ul-Hasan, P. Neutens, R. Vos, L. Lagae, P. van Dorpe, Suppression of Bulk Fluorescence Noise by Combining Waveguide-Based Near-Field Excitation and Collection, ACS Photonics 4, 495 (2017).
J. Qin, D. Zhao, S. Luo, W. Wang, J. Lu, M. Qiu, Q. Li, Strongly enhanced molecular fluorescence with ultra-thin optical magnetic mirror metasurfaces, Optics letters 42, 4478 (2017).
N. Rahbany, W. Geng, R. Bachelot, C. Couteau, Plasmon–emitter interaction using integrated ring grating–nanoantenna structures, Nanotechnology 28, 185201 (2017).
L. Zhang, A. Bluhm, K.-J. Chen, N. Larkey, S. Burrows, Performance of nano-assembly logic gates with a DNA multi-hairpin motif, Nanoscale 9, 1709 (2017).
M. Calik, T. Sick, M. Dogru, M. Döblinger, S. Datz, H. Budde, A. Hartschuh, F. Auras, T. Bein, From Highly Crystalline to Outer Surface-Functionalized Covalent Organic Frameworks-A Modulation Approach, Journal of the American Chemical Society 138, 1234 (2016).
S. Datz, C. Argyo, M. Gattner, V. Weiss, K. Brunner, J. Bretzler, C. Schirnding, A. Torrano, F. Spada, M. Vrabel, H. Engelke, C. Bräuchle, T. Carell, T. Bein, Genetically designed biomolecular capping system for mesoporous silica nanoparticles enables receptor-mediated cell uptake and controlled drug release, Nanoscale 8, 8101 (2016).
S. Krishnan, D. Ziegler, V. Arnaut, T. Martin, K. Kapsner, K. Henneberg, A. Bausch, H. Dietz, F. Simmel, Molecular transport through large-diameter DNA nanopores, Nature Communications 7, 12787 (2016).
N. Larkey, L. Zhang, S. Lansing, V. Tran, V. Seewaldt, S. Burrows, Forster resonance energy transfer to impart signal-on and -off capabilities in a single microRNA biosensor, The Analyst 141, 6239 (2016).
F. Möller, M. Kieß, D. Braun, Photochemical Microscale Electrophoresis Allows Fast Quantification of Biomolecule Binding, Journal of the American Chemical Society 138, 5363 (2016).
S. van Rijt, D. Bölükbas, C. Argyo, K. Wipplinger, M. Naureen, S. Datz, O. Eickelberg, S. Meiners, T. Bein, O. Schmid, Applicability of avidin protein coated mesoporous silica nanoparticles as drug carriers in the lung, Nanoscale 8, 8058 (2016).
B. Beliveau, A. Boettiger, M. Avendaño, R. Jungmann, R. McCole, E. Joyce, C. Kim-Kiselak, F. Bantignies, C. Fonseka, J. Erceg, Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes, Nature Communications 6 (2015).
J.-W. Janiesch, M. Weiss, G. Kannenberg, J. Hannabuss, T. Surrey, I. Platzman, J. Spatz, Key Factors for Stable Retention of Fluorophores and Labeled Biomolecules in Droplet-Based Microfluidics, Analytical Chemistry 87, 2063 (2015).
M. Li, S. Jorgensen, D. McMillan, Ł. Krzemiński, N. Daskalakis, R. Partanen, M. Tutkus, R. Tuma, D. Stamou, N. Hatzkas, L. Jeuken, Single Enzyme Experiments Reveal a Long-Lifetime Proton Leak State in a Heme-Copper Oxidase, Journal of the American Chemical Society 137, 16055 (2015).
S. Little, K. Sinsimer, J. Lee, E. Wieschaus, E. Gavis, Independent and coordinate trafficking of single Drosophila germ plasm mRNAs, Nature cell biology 17, 558 (2015).
S. Niedermayer, V. Weiss, A. Herrmann, A. Schmidt, S. Datz, K. Müller, E. Wagner, T. Bein, C. Bräuchle, Multifunctional polymer-capped mesoporous silica nanoparticles for pH-responsive targeted drug delivery, Nanoscale 7, 7953 (2015).
S. van Rijt, D. Bölükbas, C. Argyo, S. Datz, M. Lindner, O. Eickelberg, M. Konigshoff, T. Bein, S. Meiners, Protease-mediated release of chemotherapeutics from mesoporous silica nanoparticles to ex vivo human and mouse lung tumors, ACS Nano 9, 2377 (2015).
D. Porciani, G. Signore et al., Two Interconvertible Folds Modulate the Activity of a DNA Aptamer Against Transferrin Receptor, Molecular Therapy – Nucl. Acids 3, e144 (2014).
A. Tonnesen, S.M. Christensen et al., Geometrical Membrane Curvature as an Allosteric Regulator of Membrane Protein Structure and Function, Biophys. J. 106, 201 (2014).
M.P. Dobay, A. Schmidt et al., Cell Type Determines the Light-Induced Endosomal Escape Kinetics of Multifunctional Mesoporous Silica Nanoparticles, Nano Lett. 13, 1074 (2013).
J. Robert, B. Weber et al., A Three-Dimensional Engineered Artery Model for In Vitro Atherosclerosis Research, PLoS ONE 8, e79821 (2013).
L. Gogolin, H. Schroeder et al., Protein–DNA Arrays as Tools for Detection of Protein–Protein Interactions by Mass Spectrometry, ChemBioChem 14, 92 (2013).
S. Ziegler, M.E. Eberle et al., Bifunctional Oligodeoxynucleotide/AntagomiR Constructs: Evaluation of a New Tool for MicroRNA Silencing, Nucl. Acids Therapy 23, 427 (2013).
M. Fliegauf, A. F.-P. Sonnen et al., Mucociliary Clearance Defects in a Murine In Vitro Model of Pneumococcal Airway Infection, PLoS ONE 8, e59925 (2013).
M. Lehnert, C. Rosin et al., Layer-by-Layer Assembly of a Streptavidin−Fibronectin Multilayer on Biotinylated TiOX, Langmuir 29, 1732 (2013).
Y.N. Antonenko, A. Horner, P. Pohl, Electrostatically Induced Recruitment of Membrane Peptides into Clusters Requires Ligand Binding at Both Interfaces, PLoS ONE 7, e52839 (2012).
A. Kurz, M. Schwering, D.-P. Herten, Quantification of fluorescent samples by photon-antibunching, Proc. SPIE 8228, 82280K (2012).
T.D. Lazzara, D. Behn et al., Phospholipids as an alternative to direct covalent coupling: Surface functionalization of nanoporous alumina for protein recognition and purification, J. Colloid Interface Sci. 366, 57 (2012).
B. Strauss, A. Nierth et al., Direct structural analysis of modified RNA by fluorescent in-line probing, Nucl. Acids Res. 40, 861 (2012).
M. Strianese, F. De Martino et al., Myoglobin as a New Fluorescence Probe to Sense H2S, Prot. Peptide Lett. 18, 282 (2011).
N. Ehrlich, A.L. Christensen, D. Stamou, Fluorescence Anisotropy Based Single Liposome Assay to Measure Molecule-Membrane Interactions, Anal. Chem. 83, 8169 (2011).
S. Roizard, C. Danelon et al., Activation of G-Protein-Coupled Receptors in Cell-Derived Plasma Membranes Supported on Porous Beads, J. Am. Chem. Soc. 133, 16868 (2011).
W. Waldeck, G. Mueller et al., Positioning Effects of KillerRed inside of Cells correlate with DNA Strand Breaks after Activation with Visible Light, Int. J. Med. Sci. 8, 97 (2011).
R. Schmidt, J. Jacak et al., Single-Molecule Detection on a Protein-Array Assay Platform for the Exposure af a Tuberculosis Antigen, J. Proteome Res. 10, 1316 (2011).
A. Kulakowska, P. Jurkiewicz, Fluorescence Lifetime Tuning – A novel Approach to Study Flip-Flop Kinetics in Supported Phospholipid Bilayers, J. Fluoresc. 20, 563 (2010).
J. Weichsel, N. Herold et al., A Quantitative Measure for Alterations in the Actin Cytoskeleton Investigated with Automated High-Trouhput Microscopy, Cytometrie A 77, 52 (2010).
V. Cauda, H. Engelke et al., Colchine-Loaded Lipid Bilayer-Coated 50 nm Mesoporous Nanoparticles Efficiently Indude Microtubule Depolymerization upon Cell Uptake, NanoLetters 10, 2484 (2010).
V. Dinca, M. Farsari, D. Kafetzopoulos, A. Popescu, M. Dinescu et al.; Pattering parameters for biomolecules microassays constructed with nanosecond and femtosecond UV lasers, Thin Solid Films 516, 6504 (2008).
J.G. Ritter, R. Veith, J.-P. Siebrasse, U. Kubitschek, High-contrast single-particle tracking by selective focal plane illumination microscopy, Optics Express 16, 7142 (2008).
D. Wildanger, E. Rittweger, L. Kastrup, S.W. Hell, STED microscopy with a supercontinuum laser source, Optics Express 16, 9614 (2008).
A. Punge, S.O. Rizzoli et al., 3D Reconstruction of High-Resolution STED Microsopy Images, Microscopy Res. 71, 644 (2008).